Datasets for 3D genomics.
 Mota-Gomez & Rodriguez, et al 2024 METALoci Dataset cisTargetDatabase (~2Gb) and METALoci Python library available from our article."Sex-determining 3D regulatory hubs revealed by genome spatial auto-correlation analysis". by Mota-Gomez, Rodriguez, and co-authors. Submitted. (2024) |
 Molins-Pitarch, et al 2022 Geometric Diagrams of Genomes (GDG) dataset One minute video with basic principles of GDG.20' minutes video explaining the why and the how for the GDG. Model data for IMR90 cell line chr19 to reproduce its visualization. "Geometric Diagrams of Genomes: constructing a visual grammar for 3D genomics. Submitted. (2023) |
 Mugianesi, et al 2022 CHROMATIC Dataset Data (~3.7Gb) and code available for reproducing CHROMATIC results in NPC cells."CHROMATIC reveals chromatin-associated factors contributing to genome topology". by Mugianesi, Mocavini, Blanco, Di Croce and Marti-Renom. Submitted. (2022) |
 Farabella, et al 2021 Dataset Code and data (~100Mb) are available for the reproducing one of the simulations."Three-dimensional genome organization via triplex forming RNAs". by Farabella, Di Stefano, Soler-Vila, Marti-Marimon and Marti-Renom. NSMB. (2022) doi: 10.1038/s41594-021-00678-3 |
 Mendieta-Esteban, et al 2021 Dataset Code and data (~2.6Gb) are available for modeling genomes based on sparse datasets."3D reconstruction of genomic regions from sparse interaction data". by Mendieta-Esteban, Di Stefano, Castillo, Farabella and Marti-Renom. NAR Genomics and Bioinformatics 3(1) lqab017 (2021) doi: 10.1093/nargab/lqab017 |
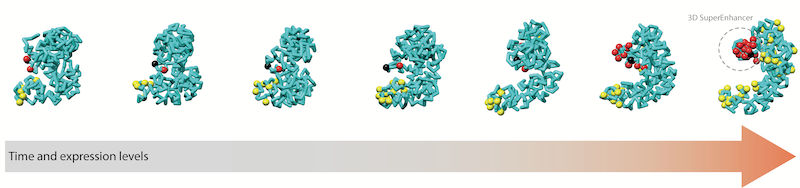 Di Stefano, et al 2020 Dataset Videos (~700Mb) are available for the 21 dynamic simulations with TADdyn.Download the code/data (~7.5Gb) to reproduce the SOX2 simulation and all figures in the manuscript: "Transcriptional activation during cell reprogramming correlates with the formation of 3D open chromatin hubs". by Di Stefano, Stadhouders, Farabella, Castillo, Serra, Graf, and Marti-Renom. Nature Communications. (2021) doi: 10.1038/s41467-020-16396-1 |
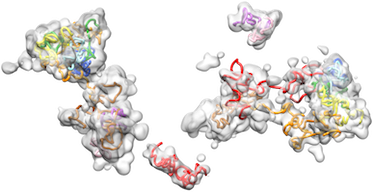 Nir, Farabella, Perez-Estrada and Ebeling et al 2018 Dataset Download a ZIP file (68Mb) that contains the OligoSTORM density maps and the IMGR models.Visit the Wu Lab Resources page for additional data on raw images and protocols described in our paper. "Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling". by Nir, Farabella, Pérez-Estrada, Ebeling, Beliveau, Sasaki, Lee, Nguyen, McCole, Chattoraj, Erceg, AlHaj-Abed, Martins, Nguyen, Hannan, Russell, Durand, Rao, Kishi, Soler-Vila, Di Pierro, Onuchic, Callahan, Schreiner, Stuckey, Yin, Lieberman-Aiden, Marti-Renom, and Wu. (2018) PLOS Genetics. 14(12):e1007872 10.1371/journal.pgen.1007872 For additional information refer to the README in the Zip file. |
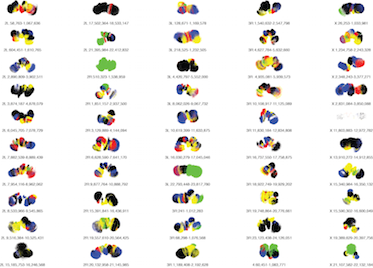 Serra et al 2015 Dataset Browse the 50 modeled regions of the fly genome described in the article:"Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors". by François Serra, Davide Baù, et al (2017) PLOS Computational Biology 13(7) e1005665. 10.1371/journal.pcbi.1005665 |
 Trussart et al 2015 Dataset Download a ZIP file (67Mb) that contains all the data described in the article:"Assessing the limits of restraint-based 3D modeling of genomes and genomic domains". by Marie Trussart, Francois Serra, Davide Baù, Ivan Junier, Luis Serrano and Marc A. Marti-Renom. NAR. (2015) doi: 10.1093/nar/gkv221 For additional information refer to the article or the README in the Zip file. |