One third of the world’s overall population is infected with Mycobacterium tuberculosis (Mtb), the causing agent of Tuberculosis (TB). About 95% of those, are thought to be in latent infection where Mtb rarely replicates. Nevertheless, ~10% of latent infections eventually progresses to active disease, which, if left untreated, kills more than half of the infected patients. Mtb infection is best described as an equilibrium involving a balance of activation and suppression of host responses, orchestrated by a complex and dynamic series of interactions between multiple host and bacterial components. Therefore, it is not unsurprising that single-target approaches for the identification of lead compounds, followed by establishment of pipelines for drug discovery, have had limited success. To address such limitation, we plan to apply genome-wide approaches to characterize the mode-of-action of a compound library, which has been validated for its activity against Mtb by GlaxoSmithKline (GSK, Partner 3), a major player in the pharmaceutical industry. Our approach uses an alternative and highly promising route by starting rather than finishing an initial process for lead compound characterization.
Aim. The GeMoA consortium, composed by four academic research groups and one company from three European countries are joining efforts with the broad aim of integrating computational and experimental approaches into an innovative platform for genome-wide characterization of the mechanism-of-action for selected chemical compounds with activity against Mtb. Our teams will combine medicinal chemistry, synthetic chemistry, computational chemistry, computational biology, genomics, transciptomics, X-ray crystallography and biochemistry to address the need for identifying new targets and compounds that can lead to unexplored new mode-of-action against Mtb. We believe that the interdisciplinary combination of such expertises will indeed add value to our research and warrant higher chances of success than any of the isolated individual efforts that form the consortium.
Work Plan. GeMoA is organized in 5 workpackages:
- WP1 “Compound selection and MedChem” aims at identifying, validating and improving selected compounds for their anti-TB activity.
- WP2 “Computational network modelling” aims at computationally building a genome-wide protein-compound network that will be used for identifying putative targets for selected compounds.
- WP3 “Genome-wide transcriptomics and target selection” aims at experimentally characterizing the effect on Mtb of a representative set of selected drugs by using gene expression profiling and genomic studies.
- WP4 “Structure and biochemistry characterization” aims at identifying the mode-of-action of selected targets and their compounds by functionally characterizing them.
- WP5 “Project coordination and managing” will aim at overseeing the overall progress of the GeMoA consortium.
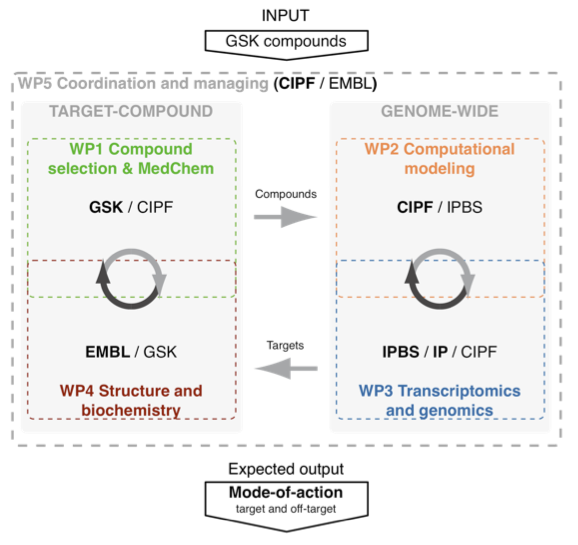
The overall pipeline of our workpackages will start by selecting a number of GSK validated compounds (WP1), which will be characterised and their targets identified using genome-wide computational modelling (WP2) and transcriptomics/genomics experiments (WP3). The identified targets will then be further validated and the mode-of-action of compounds that bind them will be characterized by structural and biochemical analysis (WP4). An iterative and integrative approach through the target-compound and the genome-wide approaches will help unravel the mode-of-action of the selected compounds. Our quantitative and interdisciplinary approach will result in rapid mode- of-action characterization via the interaction between the participating groups.
